Introduction to weather and climate data#
Key objectives and decision points
Objectives:
Understand how to work with data in the NetCDF format in your preferred language.
Understand the differences between reanalysis and interpolated data products.
Learn how to find information evaluating these data products for the variables and regions relevant to your work.
Learn how to identify major uncertainties inherent to different types of weather data products.
Decision points:
How to choose climate or weather data to use in your research project?
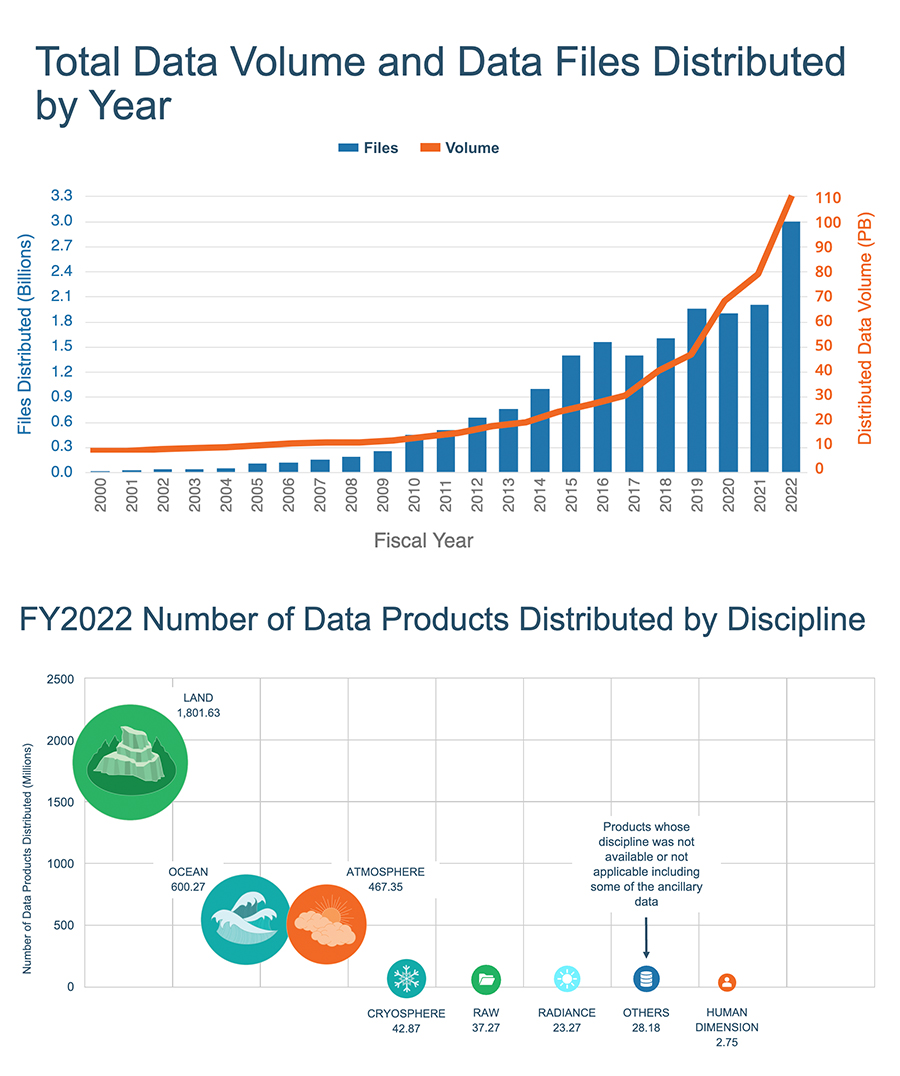
Fig. 1 The growing amount of data distributed by NASA’s EOSDIS cloud (source). We’re live through a golden age of climate data creation. See also the classic xkcd.#
This section will introduce you to the right questions to ask when deciding on climate or weather data to use in your research. It will cover how to deal with a commonly used weather and climate data format in multiple languages. It will present the differences between gridded weather data products, evaluations of weather data products, and examples of downloading several different weather data products, and a few warnings on common biases in weather data, especially precipitation.
When using weather data as independent variables in an economic model, or climate data to project your research results into the future, please keep in mind that:
There is no universally right or correct weather or climate data product.
Every weather or climate data product has its use cases, limitations, uncertainties, and quirks.
See also
Auffhammer et al. (2013) provide an excellent discussion of the challenges to consider when using weather data in econometric studies.